What is SimpleITK?
For assistance with SimpleITK at NIH, please LOGIN and return to this page
SimpleITK is an image-analysis toolkit, providing a simplified programming interface to the algorithms and data structures of the Insight Segmentation and Registration Toolkit. It supports image operations on 2D, 3D and 4D multi-channel images. The toolkit is available in eight programming languages including Python, R, Java, C#, C++, Lua, Ruby, and TCL.
SimpleITK includes several hundred classes providing a broad set of tools required for image analysis. The toolkit provides extensive support for image input and output, supporting more than 20 image file formats. A large number of filtering operations are also available, from basic image analysis operations such as edge detection, binary and grayscale morphological operations, and distance transformations to MRI bias field correction. Additionally, a variety of traditional operators used for image segmentation are available including region growing, connected component labelling, and the watershed transformation. Finally, the toolkit includes a robust registration framework, readily enabling one to align intra and inter modality images, supporting both rigid and non-rigid transformation models.
SimpleITK is widely used by researchers from various domains requiring images analysis capabilities. Amongst others these include analysis of anatomical structures imaged with CT,MR, and PET, and analysis of cellular structures imaged using dual photon microscopy, focused ion beam scanning electron microscopy and focal plane array microscopy.
The SimpleITK development team is part of the Bioinformatics and Computational Biosciences Branch under the Office of Cyber Infrastructure and Computational Biology at the National Institute of Allergy and Infectious Diseases.
If you use the toolkit in your research, show your support for its continued development by citing it in your publications. Please cite the appropriate publication:
- R. Beare, B. C. Lowekamp, Z. Yaniv, “Image Segmentation, Registration and Characterization in R with SimpleITK”, J Stat Softw, 86(8), doi: 10.18637/jss.v086.i08, 2018.
- Z. Yaniv, B. C. Lowekamp, H. J. Johnson, R. Beare, “SimpleITK Image-Analysis Notebooks: a Collaborative Environment for Education and Reproducible Research”, J Digit Imaging., doi: 10.1007/s10278-017-0037-8, 31(3): 290-303, 2018.
- B. C. Lowekamp, D. T. Chen, L. Ibáñez, D. Blezek, “The Design of SimpleITK”, Front. Neuroinform., 7:45. doi: 10.3389/fninf.2013.00045, 2013.
SimpleITK At Your Service
For assistance with SimpleITK at NIH, please LOGIN and return to this page
SimpleITK provides an extensive set of building blocks enabling development of image analysis workflows, with implementations of common workflows readily available. Short, task specific, examples are available on the SimpleITK read the docs site. Longer, more elaborate, workflows that include graphical user interfaces are available from the SimpleITK Jupyter notebooks repository. Below are the three illustrative examples.
Inter-Modal Image Registration
SimpleITK includes an interface to the ITK registration framework. One can readily register intra or inter modality images, using a variety of similarity metrics and transformation types. The registration framework is successfully used for rigid and deformable registration of anatomical and microscopy data from various sources. The images above visualize the progress of rigid registration between CT (skull) and MR (brain) images using a mutual information similarity metric. On the right, the similarity metric value minimization progresses. This case illustrates a multi-resolution framework with changes in resolution readily identified by the abrupt change in similarity metric value.
Segmentation of Microscopy Images
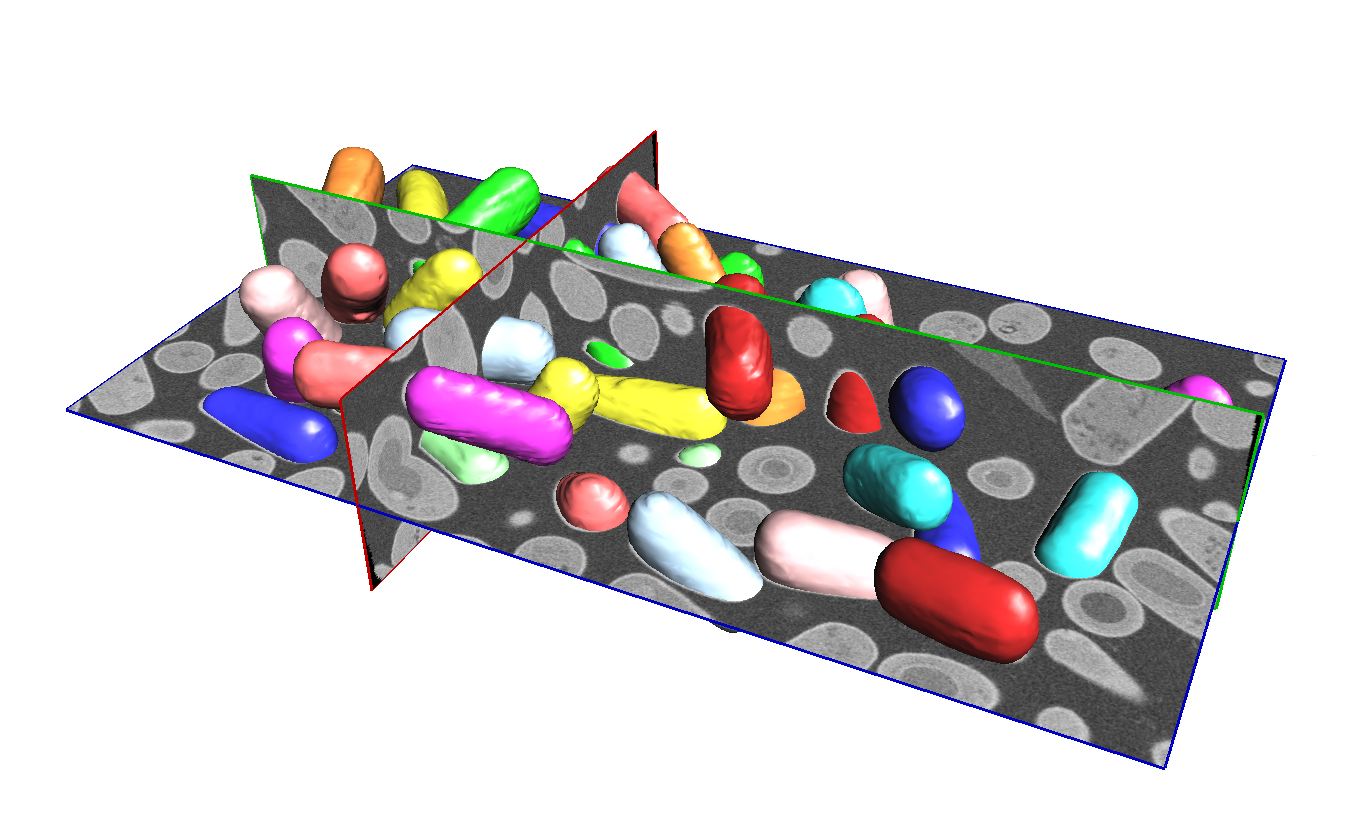
SimpleITK includes an extensive set of image processing methods to enable the development of segmentation workflows. These workflows are appropriate for novel imaging methods or unique datasets, when supervised learning, deep learning, approaches are less appropriate. Various segmentation workflows have been successfully developed for a variety of anatomical and cellular images acquired with common imaging modalities. The image above shows the results of a segmentation workflow where bacillus subtilis spores are segmented in 3D focused ion beam scanning electron microscopy. The workflow makes use of various SimpleITK filters including Otsu thresholding, distance map computation, seed based watershed computation, and various filters for analyzing the shape characteristics of the segmented bacteria.
Image Augmentation – Data Preparation for Deep Learning
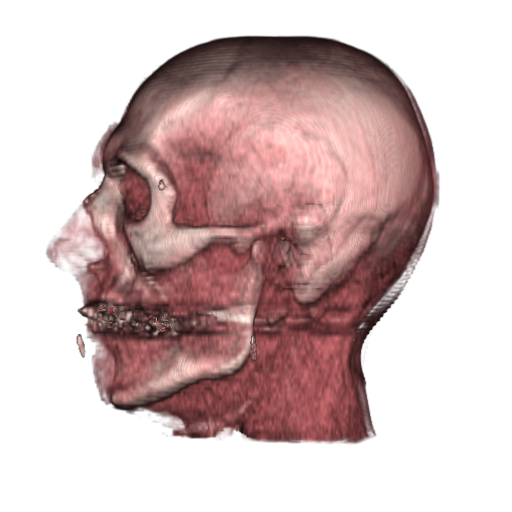

Deep learning based image analysis workflows rely on the availability of large datasets. When a dataset is not sufficiently large to capture population variability it is common to augment it by artificially introducing such variability, a process referred to as data augmentation. SimpleITK enables us to augment images using 2D and 3D transformations both affine and deformable. This capability produces more realistic spatial data augmentations with increased variability. In addition, SimpleITK provides an extensive set of intensity transformations, mimicking intensity variations that characterize various imaging devices such as bias fields in MRI. The images above show an original cranial CT volume with a size of 190x240x95 (1.01x1.01x2.5mm). The result of applying spatial data augmentation are 100 volumes with a size of 128x128x128 (1.503x1.901x1.85mm). Augmented volumes were generated using a similarity transformation, rigid + isotropic scale, with random Euler angles in [-10,10] degrees, random translations in [-10,10]mm and scaling in [0.9,1.1].
Development Team
SimpleITK is developed and managed by an interdisciplinary group of software engineers, bioinformatics specialists, analysts, and project managers from the Bioinformatics and Computational Biosciences Branch (BCBB) of the NIAID Office of Cyber Infrastructure and Computational Biology (OCICB).
- Ziv Yaniv, Ph.D.
- David Chen, Ph.D.
- Bradley Lowekamp, B.S.
For assistance with SimpleITK at NIH, please LOGIN and return to this page


