Imaging
We aim to understand the structure and distribution of components and processes at the sub-cellular, cellular, or gross anatomical levels.
For assistance with a NIAID bioinformatics or computational biology project, please LOGIN and return to this page
Examples of data analysis methods we support:
- Image classification and segmentation (Deep Learning)
- Image registration.
- Workflow development.
Imaging Team
- Ziv Yaniv, Ph.D. (Group coordinator)
- David Chen, Ph.D.
- Karthik Kantipudi, M.S.
Selected projects and publications
Microscopy Image Analysis
In collaboration with the NIAID’s Laboratory of Immune System Biology, our group has developed various extensions, both utility (channel manipulation) and algorithmic (image registration), for the Imaris (Oxford Instruments) visualization program. These extensions facilitate the creation and analysis of highly multiplexed images. This work utilizes the SimpleITK image analysis toolkit.
The code is freely available from the NIAID GitHub account: https://github.com/niaid/imaris_extensions.
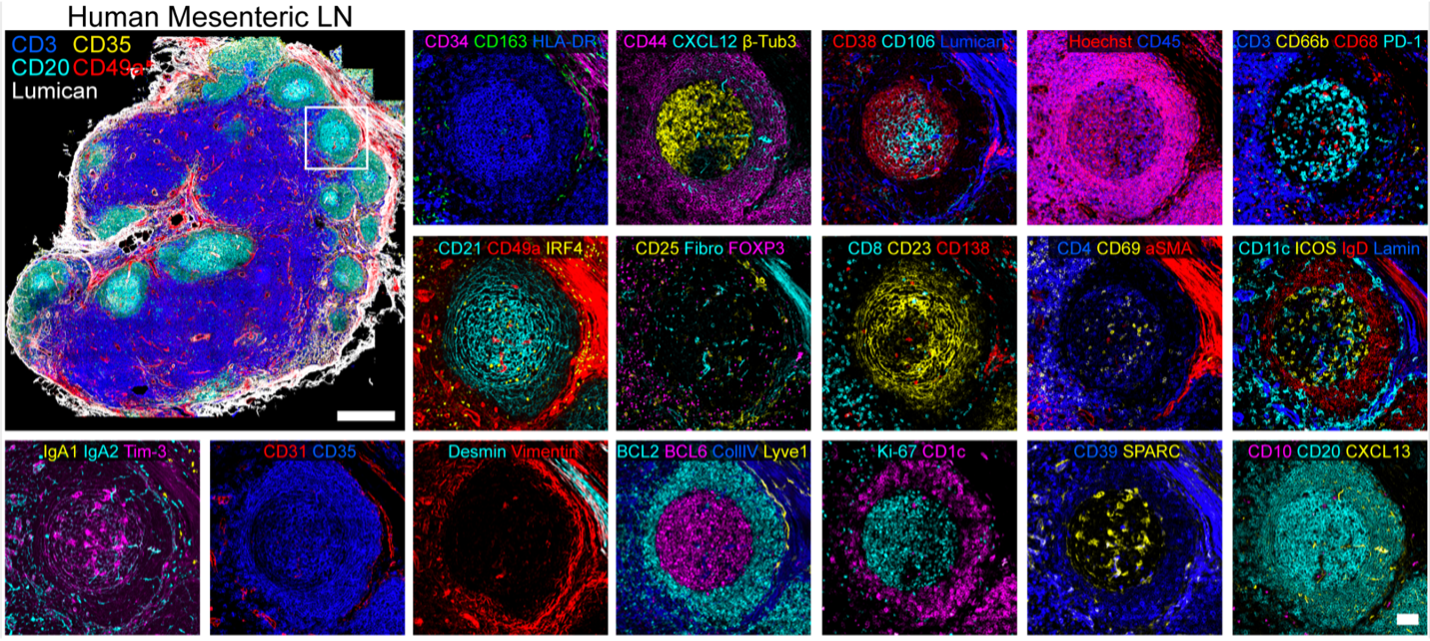
Human Mesenteric Lymph Node acquired using the IBEX protocol. Multiplex images registered using the SimpleITK based software available on GitHub.
Sample publications:
- A. J. Radtke et al., “IBEX: An iterative immunolabeling and chemical bleaching method for high-content imaging of diverse tissues”, Nat. Protocols, in press.
- A. J. Radtke et al., "IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues", Proc. Natl. Acad. Sci. USA, 117(52): 33455-33465, 2020.
Medical Image Analysis
TB Portals: As part of the TB portals program at the NIAID, our researchers are investigating the use of chest X-ray images and accompanying clinical data as a means for patient screening. This includes differentiating between TB and not-TB cases and investigating the potential for using the data to differentiate between drug-resistant and drug-sensitive TB cases. This work utilizes deep learning, relying on the keras, pytorch and MONAI deep learning frameworks.
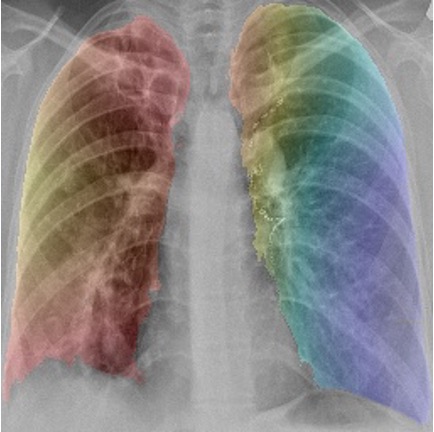
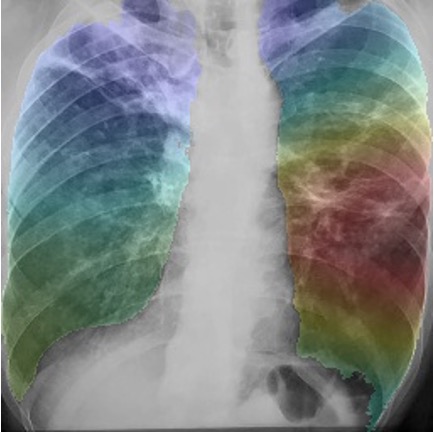
GRAD-CAM class activation maps overlaid onto images (left) drug resistant case (right) drug sensitive case.
Sample publications:
- M. Karki et al., “Identifying Drug-Resistant Tuberculosis in Chest Radiographs: Evaluation of CNN Architectures and Training Strategies”, IEEE Engineering in Medicine and Biology Conference, 2021.
- F. Yang et al., “Differentiating between drug-sensitive and drug-resistant tuberculosis with machine learning for clinical and radiological features”, Quantitative Imaging in Medicine and Surgery, in press.
Radiation exposure analysis: In collaboration with Dr. Carmen Rios, Division of Allergy, Immunology and Transplantation, NIAID, our group is developing novel visualization and image analysis methods to improve understanding of cutaneous radiation injury and countermeasures in animal models. This work utilizes the SimpleITK image analysis toolkit, the visualization toolkit VTK and 3D Slicer.
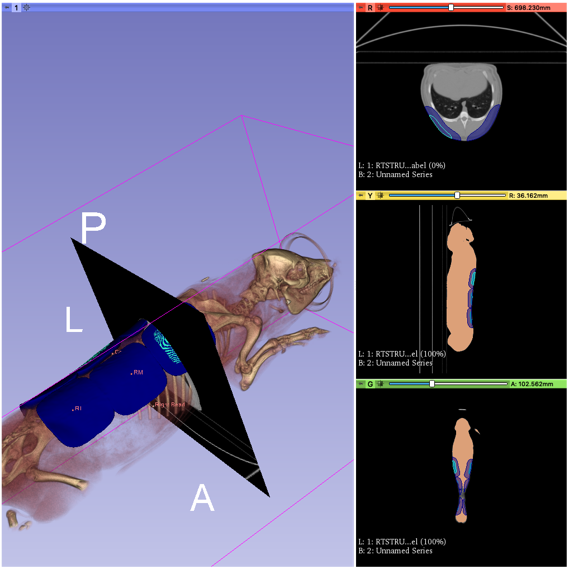
Visualization of CT scan and planned lesion locations on animal model.